Hello,
My name is Ilya Novikov, and I am a data scientist based in the Houston area. This is my home page.
For a sample of my most impactful projects please see below. Additionally, see the Skills section for a nitty-gritty overview of the tools I can use, and the About section for a quick overview of my history.
Examples of Work
- Ranking nucleotides in RNA by comparative sequence analysis
- Predicting novel protein functions by graph learning on GO similarity networks
- Analysis of Blizzard API data reveals patterns of player behavior
Ranking nucleotides in RNA by comparative sequence analysis
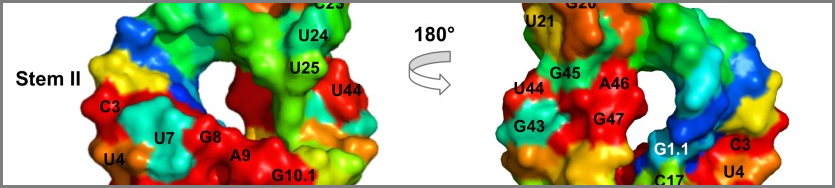
In all living cells there are molecules called Ribonucleic Acids (RNAs). Most of the them are inert templates that carry information from DNA to proteins as messenger RNA (see Central Dogma). Some RNAs, however, are actively functional - just like proteins, they are catalytic and do useful work within the cell. I helped develop a method, the RNA Evolutionary Trace, that scans the evolutionary history of these molecules and predicts their most important nucleotides. Ranking nucleotides by their importance allows wet-lab researches to narrow their efforts when studying these molecules.
The key deliverable of this project was the extensive computational validation I did to prove that the method works as advertised, published in PLOS One.
Predicting novel protein functions by graph learning on GO similarity networks
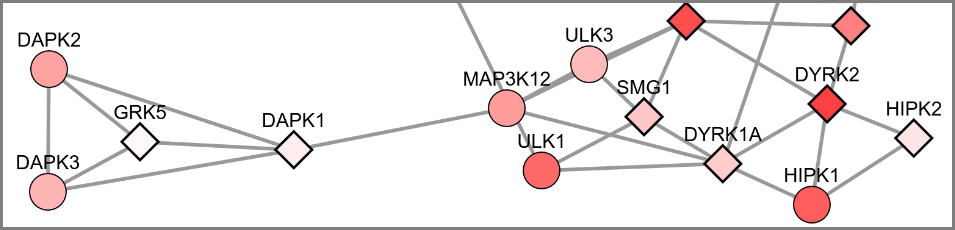 Proteins don’t work in isolation. They work together and form networks.
In practice, this means that once you’ve identified a protein associated with some
critical process, you can query that protein’s network to find additional proteins
that are involved in the same process. In case of disease, this allows you to expand
the candidate list of theraupeutic targets, and arrive at a cure faster.
Proteins don’t work in isolation. They work together and form networks.
In practice, this means that once you’ve identified a protein associated with some
critical process, you can query that protein’s network to find additional proteins
that are involved in the same process. In case of disease, this allows you to expand
the candidate list of theraupeutic targets, and arrive at a cure faster.
I developed a new approach for constructing protein networks using data from the Gene Ontology database. Gene Ontology classifies proteins with functional labels, and these labels can be used to calcualte similarity between any pair of proteins. The similarity scores can then be converted into a graph/network where most similar proteins are connected by edges. Finally, the graph itself can then be used to find clusters of related proteins using community detection or information diffusion.
The main deliverable of this project was the presentation at the International Society for Computational Biology in Prague (2017), and the web app tool linked below. Using the tool, you can diffuse labels across a GO similarity network of human kinases.
Heroku dyno that runs the app goes to sleep after 30 minutes of inactivity. So it will take 20-30 seconds for the page to respond while the dyno wakes up.
Analysis of Blizzard API data reveals patterns of player behavior
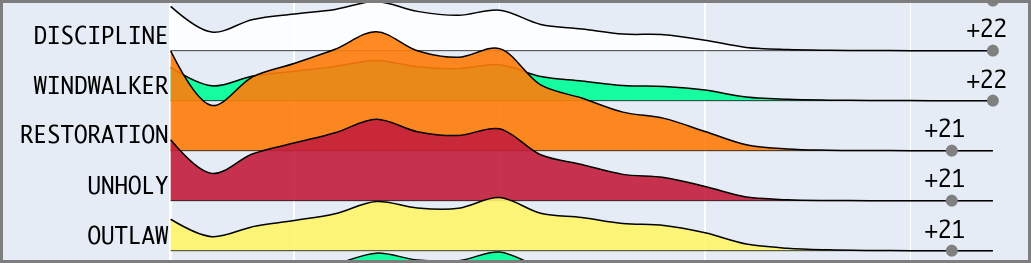 Blizzard, the operator of the World of Warcraft MMO,
provides troves of data on their player behavior. I was curious to see if I can find any
interesting patterns in that behavior. So I wrote a pipeline that continuously scans Blizzard’s API for data,
stores it in the cloud, and displays the results as this interactive dashboard.
Blizzard, the operator of the World of Warcraft MMO,
provides troves of data on their player behavior. I was curious to see if I can find any
interesting patterns in that behavior. So I wrote a pipeline that continuously scans Blizzard’s API for data,
stores it in the cloud, and displays the results as this interactive dashboard.
The dashboard allows players to see how representation of different character archetypes changes with time and level of competition. This enables players to adjust their own in-game choices to be more competitive. The dashboard is visited by 15,000 unique visitors monthly, and has been well recieved by the community. WoWHead, the game’s largest news site, did a series of articles using the dashboard as well (1, 2, 3).
The fun/instructive part of this project was dealing with large amounts of data (~150 million rows and growing), as well as building infrastructure that runs without human supervision.
